生信技能树单细胞数据挖掘笔记 (2):创建 Seurat 对象并进行质控、筛选高变基因并可视化
生信技能树单细胞数据挖掘笔记 (4):其他分析(周期判断、double 诊断、细胞类型注释)
# 3、根据 count 矩阵转换 fpkm 并完成差异分析
# 3.1 导入 count 矩阵
nm_COUNT <- read.csv("GSE81861_CRC_NM_epithelial_cells_COUNT.csv/GSE81861_CRC_NM_epithelial_cells_COUNT.csv") | |
nm_COUNT <- data_input(nm_COUNT) | |
nm_COUNT[1:4,1:4] | |
dim(nm_COUNT) | |
tumor_COUNT <- read.csv("GSE81861_CRC_tumor_epithelial_cells_COUNT.csv/GSE81861_CRC_tumor_epithelial_cells_COUNT.csv") | |
tumor_COUNT <- data_input(tumor_COUNT) | |
tumor_COUNT[1:4,1:4] | |
dim(tumor_COUNT) | |
RHC4104 RHC6087 RHL2880 RHC5949 | |
ENSG00000000003 0 428 66 141 | |
ENSG00000000005 0 0 0 0 | |
ENSG00000000419 0 179 0 1 | |
ENSG00000000457 465 0 0 0 | |
[1] 56380 160 | |
RHC4075 RHC5563 RHC5552 RHC4874 | |
ENSG00000000003 0 0 0 0 | |
ENSG00000000005 0 0 0 0 | |
ENSG00000000419 0 0 0 0 | |
ENSG00000000457 0 133 0 0 | |
[1] 56380 272 |
# 3.2 计算 fpkm 矩阵
my_FPKM <- function (counts,g_l){ | |
#### 根据有基因长度的基因,筛选矩阵子集 | |
#counts=nm_COUNT | |
ng=intersect (rownames (counts),g_l$ensembl_id) | |
length (ng) | |
lengths=g_l [match (ng,g_l$ensembl_id),2] | |
names (lengths) <- g_l [match (ng,g_l$ensembl_id),1] | |
head (lengths) | |
#### 计算样本文库大小,以及最后的 fpkm 计算 | |
counts <- counts [names (lengths),] | |
counts [1:4,1:4] | |
total_count <- colSums (counts) | |
head (total_count) | |
#根据 counts、length、total_count 计算 fpkm | |
FPKM <- t (do.call ( rbind, | |
lapply (1:length (total_count), | |
function (i){ | |
10^9*counts [,i]/lengths/total_count [i] | |
#lengths 向量自动遍历 | |
}) )) | |
FPKM [1:4,1:4] | |
return (FPKM) | |
} | |
nm_my_FPKM <- my_FPKM (counts = nm_COUNT, | |
g_l = g_l) | |
colnames (nm_my_FPKM) <- colnames (nm_COUNT) | |
nm_my_FPKM [1:4,1:4] | |
dim (nm_my_FPKM) | |
tumor_my_FPKM <- my_FPKM (counts = tumor_COUNT, | |
g_l = g_l) | |
dim (tumor_FPKM) | |
colnames (tumor_my_FPKM) <- colnames (tumor_COUNT) | |
nm_my_FPKM [1:4,1:4] | |
RHC4104 RHC6087 RHL2880 RHC5949 | |
ENSG00000000003 0.0000 160.5715 168.6839 22.7021034 | |
ENSG00000000005 0.0000 0.0000 0.0000 0.0000000 | |
ENSG00000000419 0.0000 219.5694 0.0000 0.5264304 | |
ENSG00000000457 124.2016 0.0000 0.0000 0.0000000 | |
[1] 50813 160 | |
[1] 56380 272 | |
RHC4104 RHC6087 RHL2880 RHC5949 | |
ENSG00000000003 0.0000 160.5715 168.6839 22.7021034 | |
ENSG00000000005 0.0000 0.0000 0.0000 0.0000000 | |
ENSG00000000419 0.0000 219.5694 0.0000 0.5264304 | |
ENSG00000000457 124.2016 0.0000 0.0000 0.0000000 |
# 3.3 差异分析
exp_my_FPKM <- cbind (nm_my_FPKM,tumor_my_FPKM) | |
dim (exp_my_FPKM) #转换 id 前 | |
exp_my_FPKM [1:4,1:4] #转换 id 前 | |
exp_my_FPKM <- id_change (exp_my_FPKM) | |
dim (exp_my_FPKM) #转换 id 后 | |
exp_my_FPKM [1:4,1:4] #转换 id 后 | |
my_FPKM_diff <- scRNA_deg (exp=exp_my_FPKM, group=group_dat) | |
head (my_FPKM_diff) | |
dim (my_FPKM_diff) | |
my_FPKM_diff <- my_FPKM_diff [my_FPKM_diff$p_val<0.01 & abs (my_FPKM_diff$avg_logFC)>0.8,] | |
dim (my_FPKM_diff) | |
exp_my_FPKM_diff <- exp_my_FPKM [match (rownames (my_FPKM_diff),rownames (exp_my_FPKM)),] | |
[1] 50813 432 | |
RHC4104 RHC6087 RHL2880 RHC5949 | |
ENSG00000000003 0.0000 160.5715 168.6839 22.7021034 | |
ENSG00000000005 0.0000 0.0000 0.0000 0.0000000 | |
ENSG00000000419 0.0000 219.5694 0.0000 0.5264304 | |
ENSG00000000457 124.2016 0.0000 0.0000 0.0000000 | |
[1] 50813 432 | |
[1] 24931 432 | |
[1] 24931 432 | |
RHC4104 RHC6087 RHL2880 RHC5949 | |
TSPAN6 0.0000 160.5715 168.6839 22.7021034 | |
TNMD 0.0000 0.0000 0.0000 0.0000000 | |
DPM1 0.0000 219.5694 0.0000 0.5264304 | |
SCYL3 124.2016 0.0000 0.0000 0.0000000 | |
[1] "创建 seurat 对象..." | |
[1] "质控..." | |
[1] "归一化..." | |
Performing log-normalization | |
0% 10 20 30 40 50 60 70 80 90 100% | |
[----|----|----|----|----|----|----|----|----|----| | |
**************************************************| | |
[1] "差异分析..." | |
|++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=04s | |
[1] 1231 5 | |
[1] 112 5 |
# 3.4 可视化
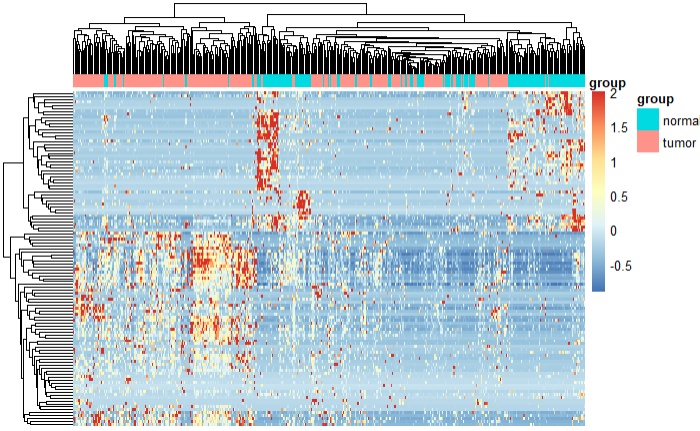
热图
my_heatmap(exp_my_FPKM_diff) |
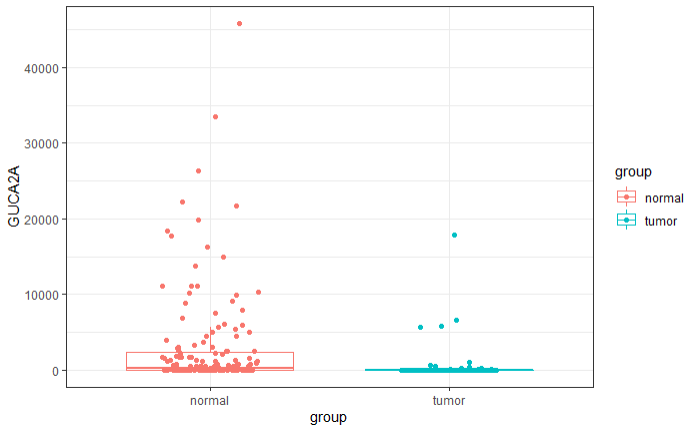
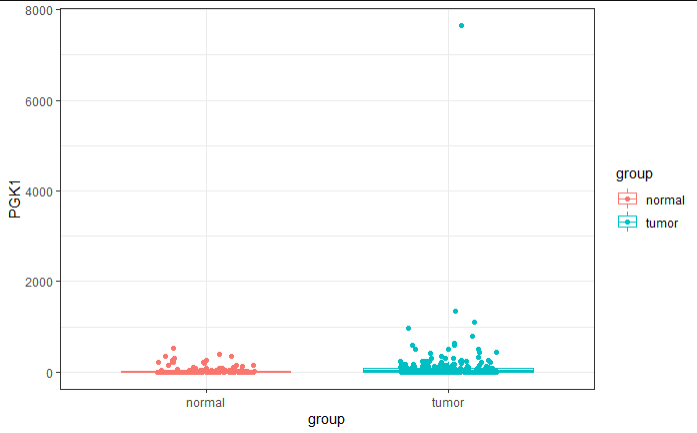
箱图